Note
Click here to download the full example code
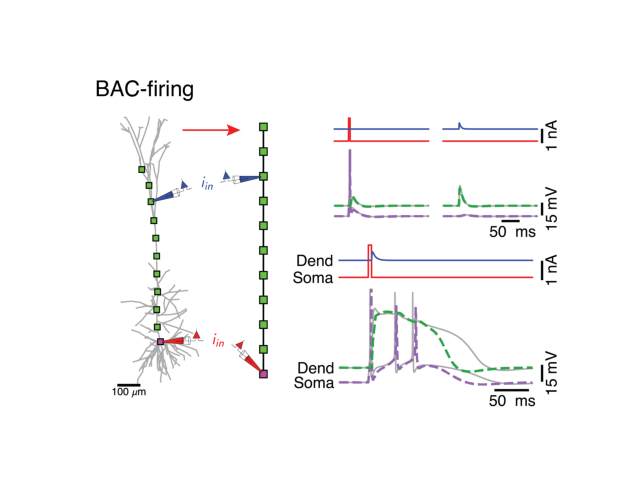
Bac firing¶

import numpy as np
import warnings
from neat import MorphLoc, CompartmentFitter
from models.L5_pyramid import getL5Pyramid
from plotutil import *
import pickle
SIM_FLAG = 1
try:
import neuron
from neat import NeuronSimTree, NeuronCompartmentTree, createReducedNeuronModel
except ImportError:
warnings.warn('NEURON not available, plotting stored image', UserWarning)
SIM_FLAG = 0
## Parameters ##################################################################
# sites for simplification
D2S_CASPIKE = np.array([685., 785., 885., 985.])
D2S_APIC = np.array([85., 185., 285., 385., 485., 585.])
CA_LOC = (224, 0.86)
# morphology color map
vals = np.ones((2, 4))
vals[0,:] = mcolors.to_rgba('DarkGrey')
vals[1,:] = mcolors.to_rgba('lime')
CMAP_MORPH = mcolors.ListedColormap(vals)
Out:
/home/docs/checkouts/readthedocs.org/user_builds/neatdend/checkouts/latest/examples/bac_firing.py:23: UserWarning: NEURON not available, plotting stored image
warnings.warn('NEURON not available, plotting stored image', UserWarning)
def getCTree(cfit, locs, f_name, recompute_ctree=False, recompute_biophys=False):
"""
Uses `neat.CompartmentFitter` to derive a `neat.CompartmentTree` for the
given `locs`. The simplified tree is stored under `f_name`. If the
simplified tree exists, it is loaded by default in memory (unless
`recompute_ctree` is ``True``). The impedances for efficient impedance
matrix evaluation are also stored, and are by default reloaded if they exist
(unless `recompute_biophys` is ``True``).
"""
try:
if recompute_ctree:
raise IOError
print('\n>>>> loading file %s'%f_name)
file = open(f_name + '.p', 'rb')
ctree = pickle.load(file)
clocs = pickle.load(file)
except (IOError, EOFError) as err:
print('\n>>>> (re-)deriving model %s'%f_name)
ctree = cfit.fitModel(locs, alpha_inds=[0], parallel=True,
use_all_channels_for_passive=False,
recompute=recompute_biophys)
clocs = ctree.getEquivalentLocs()
print('>>>> writing file %s'%f_name)
file = open(f_name + '.p', 'wb')
pickle.dump(ctree, file)
pickle.dump(clocs, file)
file.close()
return ctree, clocs
def runCaCoinc(sim_tree, locs,
ca_loc_ind, soma_ind,
stim_type='psp',
dt=0.1, t_max=300., t_calibrate=100.,
psp_params=dict(t_rise=.5, t_decay=5., i_amp=.5, t_stim=50.),
i_in_params=dict(i_amp=1.9, t_onset=45., t_dur=5.),
rec_kwargs=dict(record_from_syns=False, record_from_iclamps=False,
record_from_vclamps=False, record_from_channels=False,
record_v_deriv=False),
pprint=True):
"""
Runs the BAC-firing protocol to elicit an AP burst in response to coincident
somatic and dendritic input
"""
# initialize the NEURON model
sim_tree.initModel(dt=dt, t_calibrate=t_calibrate, factor_lambda=10.)
sim_tree.storeLocs(locs, 'rec locs')
if stim_type == 'psp' or stim_type == 'coinc':
sim_tree.addDoubleExpCurrent(locs[ca_loc_ind], psp_params['t_rise'], psp_params['t_decay'])
sim_tree.setSpikeTrain(0, psp_params['i_amp'], [psp_params['t_stim']])
if stim_type == 'psp':
sim_tree.addIClamp(locs[soma_ind], 0., i_in_params['t_onset'], i_in_params['t_dur'])
if stim_type == 'current':
sim_tree.addDoubleExpCurrent(locs[ca_loc_ind], psp_params['t_rise'], psp_params['t_decay'])
sim_tree.setSpikeTrain(0, 0., [psp_params['t_stim']])
if stim_type == 'current' or stim_type == 'coinc':
sim_tree.addIClamp(locs[soma_ind], i_in_params['i_amp'], i_in_params['t_onset'], i_in_params['t_dur'])
# simulate the NEURON model
res = sim_tree.run(t_max, pprint=pprint, **rec_kwargs)
sim_tree.deleteModel()
return res
def runCalciumCoinc(recompute_ctree=False, recompute_biophys=False, axdict=None, pshow=True):
global D2S_CASPIKE, D2S_APIC
global CA_LOC
lss_ = ['-', '-.', '--']
css_ = [colours[3], colours[0], colours[1]]
lws_ = [.8, 1.2, 1.6]
# create the full model
phys_tree = getL5Pyramid()
sim_tree = phys_tree.__copy__(new_tree=NeuronSimTree())
# compartmentfitter object
cfit = CompartmentFitter(phys_tree, name='bac_firing', path='data/')
# single branch initiation zone
branch = sim_tree.pathToRoot(sim_tree[236])[::-1]
locs_sb = sim_tree.distributeLocsOnNodes(D2S_CASPIKE, node_arg=branch, name='single branch')
# abpical trunk locations
apic = sim_tree.pathToRoot(sim_tree[221])[::-1]
locs_apic = sim_tree.distributeLocsOnNodes(D2S_APIC, node_arg=apic, name='apic connection')
# store set of locations
fit_locs = [(1, .5)] + locs_apic + locs_sb
sim_tree.storeLocs(fit_locs, name='ca coinc')
# PSP input location index
ca_ind = sim_tree.getNearestLocinds([CA_LOC], name='ca coinc')[0]
# obtain the simplified tree
ctree, clocs = getCTree(cfit, fit_locs, 'data/ctree_bac_firing',
recompute_biophys=recompute_biophys, recompute_ctree=recompute_ctree)
# print(ctree)
print('--- ctree nodes currents')
print('\n'.join([str(n.currents) for n in ctree]))
reslist, creslist_sb, creslist_sb_ = [], [], []
locindslist_sb, locindslist_apic_sb = [], []
if axdict is None:
pl.figure('inp')
axes_input = [pl.subplot(131), pl.subplot(132), pl.subplot(133)]
pl.figure('V trace')
axes_trace = [pl.subplot(131), pl.subplot(132), pl.subplot(133)]
pl.figure('morph')
axes_morph = [pl.subplot(121), pl.subplot(122)]
else:
axes_input = axdict['inp']
axes_trace = axdict['trace']
axes_morph = axdict['morph']
pshow = False
for jj, stim in enumerate(['current', 'psp', 'coinc']):
print('--- sim full ---')
rec_locs = sim_tree.getLocs('ca coinc')
# runn the simulation
res = runCaCoinc(sim_tree, rec_locs, ca_ind, 0, stim_type=stim,
rec_kwargs=dict(record_from_syns=True, record_from_iclamps=True))
print('---- sim reduced ----')
rec_locs = clocs
# run the simulation of the reduced tree
csim_tree = createReducedNeuronModel(ctree)
cres = runCaCoinc(csim_tree, rec_locs, ca_ind, 0, stim_type=stim, rec_kwargs=dict(record_from_syns=True, record_from_iclamps=True))
id_offset = 1.
vd_offset = 7.2
vlim = (-80.,20.)
ilim = (-.1,2.2)
# input current
ax = axes_input[jj]
ax.plot(res['t'], -res['i_clamp'][0], c='r', lw=lwidth)
ax.plot(res['t'], res['i_syn'][0]+id_offset, c='b', lw=lwidth)
ax.set_yticks([0., id_offset])
if jj == 1 or jj == 2:
drawScaleBars(ax, ylabel=' nA', b_offset=0)
else:
drawScaleBars(ax)
if jj == 2:
ax.set_yticklabels([r'Soma', r'Dend'])
ax.set_ylim(ilim)
# somatic trace
ax = axes_trace[jj]
ax.set_xticks([0.,50.])
ax.plot(res['t'], res['v_m'][0], c='DarkGrey', lw=lwidth)
ax.plot(cres['t'], cres['v_m'][0], c=cll[0], lw=1.6*lwidth, ls='--')
# dendritic trace
ax.plot(res['t'], res['v_m'][ca_ind]+vd_offset, c='DarkGrey', lw=lwidth)
ax.plot(cres['t'], cres['v_m'][ca_ind]+vd_offset, c=cll[1], lw=1.6*lwidth, ls='--')
ax.set_yticks([cres['v_m'][0][0], cres['v_m'][ca_ind][0]+vd_offset])
if jj == 1 or jj == 2:
drawScaleBars(ax, xlabel=' ms', ylabel=' mV', b_offset=15)
# drawScaleBars(ax, xlabel=' ms', b_offset=25)
else:
drawScaleBars(ax)
if jj == 2:
ax.set_yticklabels([r'Soma', r'Dend'])
ax.set_ylim(vlim)
print('iv')
plocs = sim_tree.getLocs('ca coinc')
markers = [{'marker': 's', 'mfc': cfl[0], 'mec': 'k', 'ms': markersize/1.1}] + \
[{'marker': 's', 'mfc': cfl[1], 'mec': 'k', 'ms': markersize/1.1} for _ in locs_apic + locs_sb]
markers[ca_ind]['marker'] = 'v'
plotargs = {'lw': lwidth/1.3, 'c': 'DarkGrey'}
sim_tree.plot2DMorphology(axes_morph[0], use_radius=False, plotargs=plotargs,
marklocs=plocs, locargs=markers, lims_margin=0.01)
# compartment tree dendrogram
labelargs = {0: {'marker': 's', 'mfc': cfl[0], 'mec': 'k', 'ms': markersize*1.2}}
labelargs.update({ii: {'marker': 's', 'mfc': cfl[1], 'mec': 'k', 'ms': markersize*1.2} for ii in range(1,len(plocs))})
ctree.plotDendrogram(axes_morph[1], plotargs={'c':'k', 'lw': lwidth}, labelargs=labelargs)
pl.show()
if __name__ == '__main__':
if SIM_FLAG:
runCalciumCoinc()
else:
plotStoredImg('../docs/figures/bac_firing.png')
Total running time of the script: ( 0 minutes 0.427 seconds)