Note
Click here to download the full example code
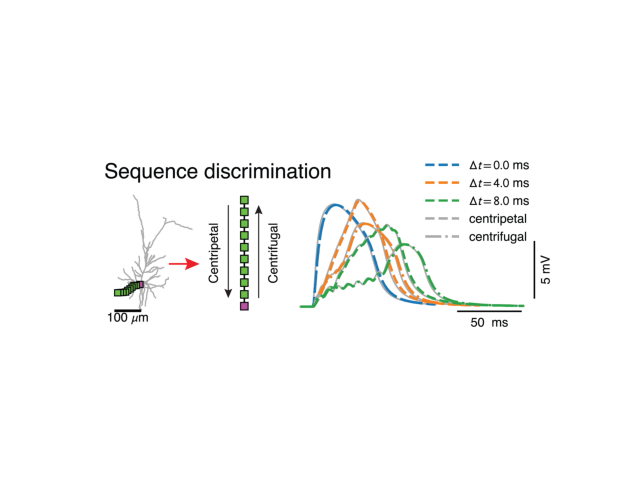
Sequence discrimination¶

import sys
import numpy as np
import warnings
from neat import MorphLoc, CompartmentFitter
from models.L23_pyramid import getL23PyramidPas
from plotutil import *
SIM_FLAG = 1
try:
import neuron
from neuron import h
from neat import NeuronSimTree, NeuronCompartmentTree, createReducedNeuronModel
except ImportError:
warnings.warn('NEURON not available, plotting stored image', UserWarning)
SIM_FLAG = 0
## Parameters ##################################################################
# synapse location parameters
SYN_NODE_IND = 112 # node index corresponding to dend[13] in Branco's (2010) model
SYN_XCOMP = [0., 0.125, 0.25, 0.375, 0.5, 0.625, 0.75, 0.875, 1.]
# AMPA synapse parameters
G_MAX_AMPA = 0.0005 # 500 pS
TAU_AMPA = 2. # ms
# NMDA synapse parameters
G_MAX_NMDA = 8000. # 8000 pS (Popen is 0.2 so effective gmax = 1600 pS, use 5000 pS for active model)
E_REV_NMDA = 5. # mV
C_MG = 1. # mM
DUR_REL = 0.5 # ms
AMP_REL = 2. # mM
Out:
/home/docs/checkouts/readthedocs.org/user_builds/neatdend/checkouts/latest/examples/sequence_discrimination.py:23: UserWarning: NEURON not available, plotting stored image
warnings.warn('NEURON not available, plotting stored image', UserWarning)
def plotSim(delta_ts=[0.,1.,2.,3.,4.,5.,6.,7.,8.], recompute=False):
class BrancoSimTree(NeuronSimTree):
'''
Inherits from :class:`NeuronSimTree` to implement Branco model
'''
def __init__(self):
super().__init__()
phys_tree = getL23PyramidPas()
phys_tree.__copy__(new_tree=self)
def setSynLocs(self):
global SYN_NODE_IND, SYN_XCOMP
# set computational tree
self.setCompTree()
self.treetype = 'computational'
# define the locations
locs = [MorphLoc((SYN_NODE_IND, x), self, set_as_comploc=True) for x in SYN_XCOMP]
self.storeLocs(locs, name='syn locs')
self.storeLocs([(1., 0.5)], name='soma loc')
# set treetype back
self.treetype = 'original'
def deleteModel(self):
super(BrancoSimTree, self).deleteModel()
self.pres = []
self.nmdas = []
def addAMPASynapse(self, loc, g_max, tau):
loc = MorphLoc(loc, self)
# create the synapse
syn = h.AlphaSynapse(self.sections[loc['node']](loc['x']))
syn.tau = tau
syn.gmax = g_max
# store the synapse
self.syns.append(syn)
def addNMDASynapse(self, loc, g_max, e_rev, c_mg, dur_rel, amp_rel):
loc = MorphLoc(loc, self)
# create the synapse
syn = h.NMDA_Mg_T(self.sections[loc['node']](loc['x']))
syn.gmax = g_max
syn.Erev = e_rev
syn.mg = c_mg
# create the presynaptic segment for release
pre = h.Section(name='pre %d'%len(self.pres))
pre.insert('release_BMK')
pre(0.5).release_BMK.dur = dur_rel
pre(0.5).release_BMK.amp = amp_rel
# connect
h.setpointer(pre(0.5).release_BMK._ref_T, 'C', syn)
# store the synapse
self.nmdas.append(syn)
self.pres.append(pre)
# setpointer cNMDA[n].C, PRE[n].T_rel(0.5)
# setpointer im_xtra(x), i_membrane(x)
# h.setpointer(dend(seg.x)._ref_i_membrane, 'im', dend(seg.x).xtra)
def setSpikeTime(self, syn_index, spike_time):
spk_tm = spike_time + self.t_calibrate
# add spike for AMPA synapse
self.syns[syn_index].onset = spk_tm
# add spike for NMDA synapse
self.pres[syn_index](0.5).release_BMK.delay = spk_tm
def addAllSynapses(self):
global G_MAX_AMPA, TAU_AMPA, G_MAX_NMDA, E_REV_NMDA, C_MG, DUR_REL, AMP_REL
for loc in self.getLocs('syn locs'):
# ampa synapse
self.addAMPASynapse(loc, G_MAX_AMPA, TAU_AMPA)
# nmda synapse
self.addNMDASynapse(loc, G_MAX_NMDA, E_REV_NMDA, C_MG, DUR_REL, AMP_REL)
def setSequence(self, delta_t, centri='fugal', t0=10., tadd=100.):
n_loc = len(self.getLocs('syn locs'))
if centri == 'fugal':
for ll in range(n_loc):
self.setSpikeTime(ll, t0 + ll * delta_t)
elif centri == 'petal':
for tt, ll in enumerate(range(n_loc)[::-1]):
self.setSpikeTime(ll, t0 + tt * delta_t)
else:
raise IOError('Only centrifugal or centripetal sequences are allowed, ' + \
'use \'fugal\' resp. \'petal\' as second arg.')
return n_loc * delta_t + t0 + tadd
def reduceModel(self, pprint=False):
global SYN_NODE_IND, SYN_XCOMP
locs = [MorphLoc((1, .5), self, set_as_comploc=True)] + \
[MorphLoc((SYN_NODE_IND, x), self, set_as_comploc=True) for x in SYN_XCOMP]
# creat the reduced compartment tree
ctree = self.createCompartmentTree(locs)
# create trees to derive fitting matrices
sov_tree, greens_tree = self.getZTrees()
# compute the steady state impedance matrix
z_mat = greens_tree.calcImpedanceMatrix(locs)[0].real
# fit the conductances to steady state impedance matrix
ctree.computeGMC(z_mat, channel_names=['L'])
if pprint:
np.set_printoptions(precision=1, linewidth=200)
print(('Zmat original (MOhm) =\n' + str(z_mat)))
print(('Zmat fitted (MOhm) =\n' + str(ctree.calcImpedanceMatrix())))
# get SOV constants
alphas, phimat = sov_tree.getImportantModes(locarg=locs,
sort_type='importance', eps=1e-12)
# n_mode = len(locs)
# alphas, phimat = alphas[:n_mode], phimat[:n_mode, :]
importance = sov_tree.getModeImportance(sov_data=(alphas, phimat), importance_type='full')
# fit the capacitances from SOV time-scales
# ctree.computeC(-alphas*1e3, phimat, weight=importance)
ctree.computeC(-alphas[:1]*1e3, phimat[:1,:], importance=importance[:1])
if pprint:
print(('Taus original (ms) =\n' + str(np.abs(1./alphas))))
lambdas, _, _ = ctree.calcEigenvalues()
print(('Taus fitted (ms) =\n' + str(np.abs(1./lambdas))))
return ctree
def runSim(self, delta_t=12.):
try:
el = self[0].currents['L'][1]
except AttributeError:
el = self[1].currents['L'][1]
# el=-75.
self.initModel(dt=0.025, t_calibrate=0., v_init=el, factor_lambda=10.)
# add the synapses
self.addAllSynapses()
t_max = self.setSequence(delta_t, centri='petal')
# set recording locs
self.storeLocs(self.getLocs('soma loc') + self.getLocs('syn locs'), name='rec locs')
# run the simulation
res_centripetal = self.run(t_max, pprint=True)
# delete the model
self.deleteModel()
self.initModel(dt=0.025, t_calibrate=0., v_init=el, factor_lambda=10.)
# add the synapses
self.addAllSynapses()
t_max = self.setSequence(delta_t, centri='fugal')
# set recording locs
self.storeLocs(self.getLocs('soma loc') + self.getLocs('syn locs'), name='rec locs')
# run the simulation
res_centrifugal = self.run(t_max, pprint=True)
# delete the model
self.deleteModel()
return res_centripetal, res_centrifugal
class BrancoReducedTree(NeuronCompartmentTree, BrancoSimTree):
def __init__(self):
# call the initializer of :class:`NeuronSimTree`, follows after
# :class:`BrancoSimTree` in MRO
super(BrancoSimTree, self).__init__(file_n=None, types=[1,3,4])
def setSynLocs(self, equivalent_locs):
self.storeLocs(equivalent_locs[1:], name='syn locs')
self.storeLocs(equivalent_locs[:1], name='soma loc')
global SYN_NODE_IND, SYN_XCOMP
# initialize the full model
simtree = BrancoSimTree()
simtree.setSynLocs()
simtree.setCompTree()
# derive the reduced model retaining only soma and synapse locations
fit_locs = simtree.getLocs('soma loc') + simtree.getLocs('syn locs')
c_fit = CompartmentFitter(simtree, name='sequence_discrimination', path='data/')
ctree = c_fit.fitModel(fit_locs, recompute=recompute)
clocs = ctree.getEquivalentLocs()
# create the reduced model for NEURON simulation
csimtree_ = createReducedNeuronModel(ctree)
csimtree = csimtree_.__copy__(new_tree=BrancoReducedTree())
csimtree.setSynLocs(clocs)
pl.figure('Branco', figsize=(5,5))
gs = GridSpec(2,2)
ax0 = pl.subplot(gs[0,0])
ax_ = pl.subplot(gs[0,1])
ax1 = myAx(pl.subplot(gs[1,:]))
# plot the full morphology
locargs = [dict(marker='s', mec='k', mfc=cfl[0], ms=markersize)]
locargs.extend([dict(marker='s', mec='k', mfc=cfl[1], ms=markersize) for ii in range(1,len(fit_locs))])
pnodes = [n for n in simtree if n.swc_type != 2]
plotargs = {'lw': lwidth/1.3, 'c': 'DarkGrey'}
simtree.plot2DMorphology(ax0, use_radius=False,node_arg=pnodes,
plotargs=plotargs, marklocs=fit_locs, locargs=locargs, lims_margin=.01,
textargs={'fontsize': ticksize}, labelargs={'fontsize': ticksize})
# plot a schematic of the reduced model
labelargs = {0: {'marker': 's', 'mfc': cfl[0], 'mec': 'k', 'ms': markersize*1.2}}
labelargs.update({ii: {'marker': 's', 'mfc': cfl[1], 'mec': 'k', 'ms': markersize*1.2} for ii in range(1,len(fit_locs))})
ctree.plotDendrogram(ax_, plotargs={'c':'k', 'lw': lwidth}, labelargs=labelargs)
xlim, ylim = np.array(ax_.get_xlim()), np.array(ax_.get_ylim())
pp = np.array([np.mean(xlim), np.mean(ylim)])
dp = np.array([2.*np.abs(xlim[1]-xlim[0])/3.,0.])
ax_.annotate('Centrifugal', #xycoords='axes points',
xy=pp, xytext=pp+dp,
size=ticksize, rotation=90, ha='center', va='center')
ax_.annotate('Centripetal', #xycoords='axes points',
xy=pp, xytext=pp-dp,
size=ticksize, rotation=90, ha='center', va='center')
# plot voltage traces
legend_handles = []
for ii, delta_t in enumerate(delta_ts):
res_cp, res_cf = simtree.runSim(delta_t=delta_t)
ax1.plot(res_cp['t'], res_cp['v_m'][0], c='DarkGrey', lw=lwidth)
ax1.plot(res_cf['t'], res_cf['v_m'][0], c='DarkGrey', lw=lwidth)
cres_cp, cres_cf = csimtree.runSim(delta_t=delta_t)
line = ax1.plot(cres_cp['t'], cres_cp['v_m'][0], c=colours[ii%len(colours)],
ls='--', lw=1.6*lwidth, label=r'$\Delta t = ' + '%.1f$ ms'%delta_t)
ax1.plot(cres_cf['t'], cres_cf['v_m'][0], c=colours[ii%len(colours)], ls='-.', lw=1.6*lwidth)
legend_handles.append(line[0])
legend_handles.append(mlines.Line2D([0,0],[0,0], ls='--', lw=1.6*lwidth, c='DarkGrey', label=r'centripetal'))
legend_handles.append(mlines.Line2D([0,0],[0,0], ls='-.', lw=1.6*lwidth, c='DarkGrey', label=r'centrifugal'))
drawScaleBars(ax1, r' ms', r' mV', b_offset=20, h_offset=15, v_offset=15)
myLegend(ax1, loc='upper left', bbox_to_anchor=(.7,1.3), handles=legend_handles,
fontsize=ticksize, handlelength=2.7)
pl.tight_layout()
pl.show()
if __name__ == '__main__':
if SIM_FLAG:
plotSim(delta_ts=[0.,4.,8.])
else:
plotStoredImg('../docs/figures/sequence_discrimination.png')
Total running time of the script: ( 0 minutes 0.206 seconds)