Note
Click here to download the full example code
Basal AP Backprop¶

import numpy as np
import warnings
from neat import MorphLoc, CompartmentFitter
from models.L23_pyramid import getL23PyramidNaK
from plotutil import *
import pickle
SIM_FLAG = 1
try:
import neuron
from neat import NeuronSimTree, NeuronCompartmentTree, createReducedNeuronModel
except ImportError:
warnings.warn('NEURON not available, plotting stored image', UserWarning)
SIM_FLAG = 0
## Parameters ##################################################################
# soma nodes branco
SLOCS = [(1, .5)]
# loc params
D2S_BASAL = np.array([50., 100., 150.])
# soma stimulus params
STIM_PARAMS = {'amp': 3., # nA
't_onset': 5., # ms
't_dur': 1. # ms
}
# simulation parameters
DT = 0.025
T_MAX = 300.
TC = 200.
# morphology color map
vals = np.ones((2, 4))
vals[0,:] = mcolors.to_rgba('DarkGrey')
vals[1,:] = mcolors.to_rgba('lime')
CMAP_MORPH = mcolors.ListedColormap(vals)
Out:
/home/docs/checkouts/readthedocs.org/user_builds/neatdend/checkouts/latest/examples/basal_ap_backprop.py:23: UserWarning: NEURON not available, plotting stored image
warnings.warn('NEURON not available, plotting stored image', UserWarning)
def getCTree(cfit, locs, f_name, recompute_ctree=False, recompute_biophys=False):
"""
Uses `neat.CompartmentFitter` to derive a `neat.CompartmentTree` for the
given `locs`. The simplified tree is stored under `f_name`. If the
simplified tree exists, it is loaded by default in memory (unless
`recompute_ctree` is ``True``). The impedances for efficient impedance
matrix evaluation are also stored, and are by default reloaded if they exist
(unless `recompute_biophys` is ``True``).
"""
try:
if recompute_ctree:
raise IOError
print('\n>>>> loading file %s'%f_name)
file = open(f_name + '.p', 'rb')
ctree = pickle.load(file)
clocs = pickle.load(file)
except (IOError, EOFError) as err:
print('\n>>>> (re-)deriving model %s'%f_name)
ctree = cfit.fitModel(locs, alpha_inds=[0], parallel=True,
use_all_channels_for_passive=False,
recompute=recompute_biophys)
clocs = ctree.getEquivalentLocs()
print('>>>> writing file %s'%f_name)
file = open(f_name + '.p', 'wb')
pickle.dump(ctree, file)
pickle.dump(clocs, file)
file.close()
return ctree, clocs
def calcAmpDelayWidth(res):
"""
Compute a number of AP amplitude, delay compared to start of simulation,
delay of backpropagating AP compared to soma AP, and halfwidth
"""
dt = res['t'][1] - res['t'][0]
# amplitude of peak
res['amp'] = np.max(res['v_m'], axis=1) - res['v_m'][:,0]
# delay of peak compared to soma
res['delay'] = dt * (np.argmax(res['v_m'], axis=1) - np.argmax(res['v_m'][0]))
# absolute delay of peak
res['dop'] = dt * np.argmax(res['v_m'], axis=1)
# width of waveform at half amplitude
v_half = res['amp'] / 2. + res['v_m'][:,0]
res['width'] = dt*np.sum(res['v_m'] > v_half[:,None], axis=1)
def runSim(simtree, locs, soma_loc, stim_params={'amp':.5, 't_onset':5., 't_dur':1.}):
"""
Runs simulation to inject somatic current in order to elicit AP
"""
global DT, T_MAX, TC
global T_DUR, G_SYN, N_INP
simtree.initModel(dt=DT, t_calibrate=TC, factor_lambda=1.)
simtree.addIClamp(soma_loc, stim_params['amp'], stim_params['t_onset'], stim_params['t_dur'])
simtree.storeLocs([soma_loc] + locs, 'rec locs')
res = simtree.run(40., record_from_iclamps=True)
simtree.deleteModel()
return res
def basalAPBackProp(recompute_ctree=False, recompute_biophys=False, axes=None, pshow=True):
global STIM_PARAMS, D2S_BASAL, SLOCS
global CMAP_MORPH
rc, rb = recompute_ctree, recompute_biophys
if axes is None:
pl.figure(figsize=(7,5))
ax1, ax2, ax4, ax5 = pl.subplot(221), pl.subplot(223), pl.subplot(222), pl.subplot(224)
divider = make_axes_locatable(ax1)
ax3 = divider.append_axes("top", "30%", pad="10%")
ax4, ax5 = myAx(ax4), myAx(ax5)
pl.figure(figsize=(5,5))
gs = GridSpec(2,2)
ax_morph, ax_red1, ax_red2 = pl.subplot(gs[0,:]), pl.subplot(gs[1,0]), pl.subplot(gs[1,1])
else:
ax1, ax2, ax3 = axes['trace']
ax4, ax5 = axes['amp-delay']
ax_morph, ax_red1, ax_red2 = axes['morph']
pshow = False
# create the full model
phys_tree = getL23PyramidNaK()
sim_tree = phys_tree.__copy__(new_tree=NeuronSimTree())
# distribute locations to measure backAPs on branches
leafs_basal = [node for node in sim_tree.leafs if node.swc_type == 3]
branches = [sim_tree.pathToRoot(leaf)[::-1] for leaf in leafs_basal]
locslist = [sim_tree.distributeLocsOnNodes(D2S_BASAL, node_arg=branch) for branch in branches]
branchlist = [b for ii, b in enumerate(branches) if len(locslist[ii]) == 3]
locs = [locs for locs in locslist if len(locs) == 3][1]
# do back prop sims
amp_diffs_3loc, delay_diffs_3loc = np.zeros(3), np.zeros(3)
amp_diffs_1loc, delay_diffs_1loc = np.zeros(3), np.zeros(3)
amp_diffs_biop, delay_diffs_biop = np.zeros(3), np.zeros(3)
# compartmentfitter object
cfit = CompartmentFitter(phys_tree, name='basal_bAP', path='data/')
# create reduced tree
ctree, clocs = getCTree(cfit, [SLOCS[0]] + locs, 'data/ctree_basal_bAP_3loc',
recompute_ctree=rc, recompute_biophys=rb)
csimtree = createReducedNeuronModel(ctree)
print(ctree)
# run the simulation of he full tree
res = runSim(sim_tree, locs, SLOCS[0], stim_params=STIM_PARAMS)
calcAmpDelayWidth(res)
amp_diffs_biop[:] = res['amp'][1:]
delay_diffs_biop[:] = res['delay'][1:]
# run the simulation of the reduced tree
cres = runSim(csimtree, clocs[1:], clocs[0], stim_params=STIM_PARAMS)
calcAmpDelayWidth(cres)
amp_diffs_3loc[:] = cres['amp'][1:]
delay_diffs_3loc[:] = cres['delay'][1:]
# reduced models with one single dendritic site
creslist = []
for jj, loc in enumerate(locs):
# create reduced tree with all 1 single dendritic site locs
ctree, clocs = getCTree(cfit, [SLOCS[0]] + [loc], 'data/ctree_basal_bAP_1loc%d'%jj,
recompute_ctree=rc, recompute_biophys=False)
csimtree = createReducedNeuronModel(ctree)
print(ctree)
# run the simulation of the reduced tree
cres_ss = runSim(csimtree, [clocs[1]], clocs[0], stim_params=STIM_PARAMS)
calcAmpDelayWidth(cres_ss)
creslist.append(cres_ss)
amp_diffs_1loc[jj] = cres_ss['amp'][1]
delay_diffs_1loc[jj] = cres_ss['delay'][1]
ylim = (-90., 60.)
x_range = np.array([-3.,14])
xlim = (0., 12.)
tp_full = res['t'][np.argmax(res['v_m'][0])]
tp_3comp = cres['t'][np.argmax(cres['v_m'][0])]
tp_1comp = creslist[2]['t'][np.argmax(creslist[2]['v_m'][0])]
tlim_full = tp_full + x_range
tlim_3comp = tp_3comp + x_range
tlim_1comp = tp_1comp + x_range
i0_full, i1_full = np.round(tlim_full / DT).astype(int)
i0_3comp, i1_3comp = np.round(tlim_3comp / DT).astype(int)
i0_1comp, i1_1comp = np.round(tlim_1comp / DT).astype(int)
ax1.set_ylabel(r'soma')
ax1.plot(res['t'][i0_full:i1_full] - tlim_full[0], res['v_m'][0][i0_full:i1_full],
lw=lwidth, c='DarkGrey', label=r'full')
ax1.plot(cres['t'][i0_3comp:i1_3comp] - tlim_3comp[0], cres['v_m'][0][i0_3comp:i1_3comp],
ls='--', lw=1.6*lwidth, c=colours[0], label=r'3 comp')
ax1.plot(creslist[2]['t'][i0_1comp:i1_1comp] - tlim_1comp[0], creslist[2]['v_m'][0][i0_1comp:i1_1comp],
ls='-.', lw=1.6*lwidth, c=colours[1], label=r'1 comp')
ax1.set_ylim(ylim)
# ax1.set_xlim(xlim)
drawScaleBars(ax1, b_offset=15)
myLegend(ax1, add_frame=False, loc='center left', bbox_to_anchor=[0.35, 0.55], fontsize=ticksize,
labelspacing=.8, handlelength=2., handletextpad=.2)
ax2.set_ylabel(r'dend' + '\n($d_{soma} = 150$ $\mu$m)')
ax2.plot(res['t'][i0_full:i1_full] - tlim_full[0], res['v_m'][3][i0_full:i1_full],
lw=lwidth, c='DarkGrey', label=r'full')
ax2.plot(cres['t'][i0_3comp:i1_3comp] - tlim_3comp[0], cres['v_m'][3][i0_3comp:i1_3comp],
ls='--', lw=1.6*lwidth, c=colours[0], label=r'3 comp')
ax2.plot(creslist[2]['t'][i0_1comp:i1_1comp] - tlim_1comp[0], creslist[2]['v_m'][1][i0_1comp:i1_1comp],
ls='-.', lw=1.6*lwidth, c=colours[1], label=r'1 comp')
imax = np.argmax(res['v_m'][3])
xp = res['t'][imax]
ax2.annotate(r'$v_{amp}$',
xy=(xlim[0], np.mean(ylim)), xytext=(xlim[0], np.mean(ylim)),
fontsize=ticksize, ha='center', va='center', rotation=90.)
ax2.annotate(r'$t_{delay}$',
xy=(xp, ylim[1]), xytext=(xp, ylim[1]),
fontsize=ticksize, ha='center', va='center', rotation=0.)
ax2.set_ylim(ylim)
ax2.set_xlim(xlim)
drawScaleBars(ax2, xlabel=' ms', ylabel=' mV', b_offset=15)
# myLegend(ax2, add_frame=False, ncol=2, fontsize=ticksize,
# loc='upper center', bbox_to_anchor=[.5, -.1],
# labelspacing=.6, handlelength=2., handletextpad=.2, columnspacing=.5)
ax3.plot(res['t'][i0_full:i1_full] - tlim_full[0], -res['i_clamp'][0][i0_full:i1_full],
lw=lwidth, c='r')
ax3.set_yticks([0.,3.])
drawScaleBars(ax3, ylabel=' nA', b_offset=0)
# ax3.set_xlim(xlim)
# color the branches
cnodes = [b for branch in branches for b in branch]
if cnodes is None:
plotargs = {'lw': lwidth/1.3, 'c': 'DarkGrey'}
cs = {node.index: 0 for node in sim_tree}
else:
plotargs = {'lw': lwidth/1.3}
cinds = [n.index for n in cnodes]
cs = {node.index: 1 if node.index in cinds else 0 for node in sim_tree}
# mark example locations
plocs = [SLOCS[0]] + locs
markers = [{'marker': 's', 'c': cfl[0], 'mec': 'k', 'ms': markersize}] + \
[{'marker': 's', 'c': cfl[1], 'mec': 'k', 'ms': markersize} for _ in plocs[1:]]
# plot morphology
sim_tree.plot2DMorphology(ax_morph, use_radius=False, plotargs=plotargs,
cs=cs, cmap=CMAP_MORPH,
marklocs=plocs, locargs=markers, lims_margin=0.01)
# plot compartment tree schematic
ctree_3l = cfit.setCTree([SLOCS[0]] + locs)
ctree_3l = cfit.ctree
ctree_1l = cfit.setCTree([SLOCS[0]] + locs[0:1])
ctree_1l = cfit.ctree
labelargs = {0: {'marker': 's', 'mfc': cfl[0], 'mec': 'k', 'ms': markersize*1.2}}
labelargs.update({ii: {'marker': 's', 'mfc': cfl[1], 'mec': 'k', 'ms': markersize*1.2} for ii in range(1,len(plocs))})
ctree_3l.plotDendrogram(ax_red1, plotargs={'c':'k', 'lw': lwidth}, labelargs=labelargs)
labelargs = {0: {'marker': 's', 'mfc': cfl[0], 'mec': 'k', 'ms': markersize*1.2},
1: {'marker': 's', 'mfc': cfl[1], 'mec': 'k', 'ms': markersize*1.2}}
ctree_1l.plotDendrogram(ax_red2, plotargs={'c':'k', 'lw': lwidth}, labelargs=labelargs)
ax_red1.set_xticks([]); ax_red1.set_yticks([])
ax_red1.set_xlabel(r'$\Delta x = 50$ $\mu$m', fontsize=ticksize,rotation=60)
ax_red2.set_xticks([]); ax_red2.set_yticks([])
ax_red2.set_xlabel(r'$\Delta x = 150$ $\mu$m', fontsize=ticksize,rotation=60)
xb = np.arange(3)
bwidth = 1./4.
xtls = [r'50', r'100', r'150']
ax4, ax5 = myAx(ax4), myAx(ax5)
ax4.bar(xb-bwidth, amp_diffs_biop, width=bwidth, align='center', color='DarkGrey', edgecolor='k', label=r'full')
ax4.bar(xb, amp_diffs_3loc, width=bwidth, align='center', color=colours[0], edgecolor='k', label=r'4 comp')
ax4.bar((xb+bwidth)[-1:], amp_diffs_1loc[-1:], width=bwidth, align='center', color=colours[1], edgecolor='k', label=r'2 comp')
ax4.set_ylabel(r'$v_{amp}$ (mV)')
ax4.set_xticks(xb)
ax4.set_xticklabels([])
ax4.set_ylim(50.,110.)
ax4.set_yticks([50., 80.])
myLegend(ax4, add_frame=False, loc='lower center', bbox_to_anchor=[.5, 1.05], fontsize=ticksize,
labelspacing=.1, handlelength=1., handletextpad=.2, columnspacing=.5)
ax5.bar(xb-bwidth, delay_diffs_biop, width=bwidth, align='center', color='DarkGrey', edgecolor='k', label=r'full')
ax5.bar(xb, delay_diffs_3loc, width=bwidth, align='center', color=colours[0], edgecolor='k', label=r'4 comp')
ax5.bar((xb+bwidth)[-1:], delay_diffs_1loc[-1:], width=bwidth, align='center', color=colours[1], edgecolor='k', label=r'2 comp')
ax5.set_ylabel(r'$t_{delay}$ (ms)')
ax5.set_xticks(xb)
ax5.set_xticklabels(xtls)
ax5.set_xlabel(r'$d_{soma}$ ($\mu$m)')
ax5.set_yticks([0., 0.5])
if pshow:
pl.show()
if __name__ == '__main__':
if SIM_FLAG:
basalAPBackProp()
else:
plotStoredImg('../docs/figures/ap_backpropagation.png')
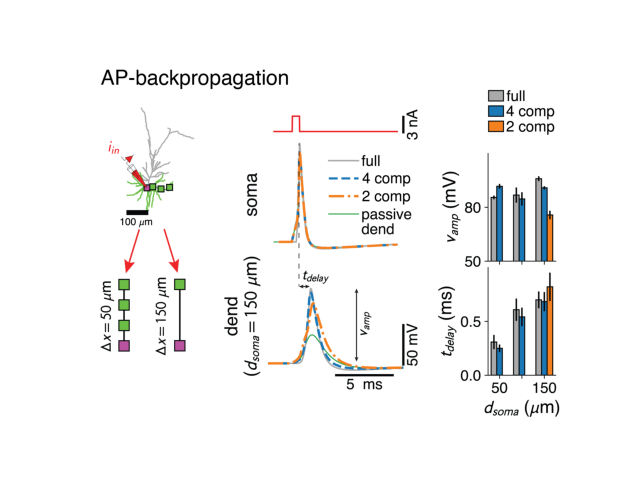
Total running time of the script: ( 0 minutes 0.404 seconds)